Nuclear Magnetic Resonance (NMR) is widely employed for protein structure determination and measuring protein-target interactions. In our studies, two dimensional heteronuclear single quantum correlation (HSQC) experiments are employed to study the interactions of proteins with ion exchange and multimodal (MM) chromatographic ligands in solution.
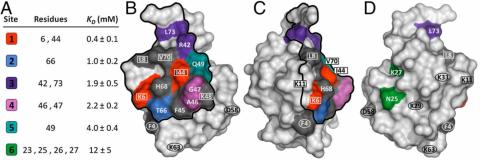
This technique makes it possible to identify the ligand interaction sites on the protein surface and apparent binding affinities for each amino acid residue.

This technique was also employed to evaluate the effect of fluid phase modifier (e.g. Urea) on protein binding in MM chromatographic systems. The results showed that, while the binding region of the protein was preserved in presence of urea, the strength of binding decreased due to weak interactions of certain residues in the binding patch.
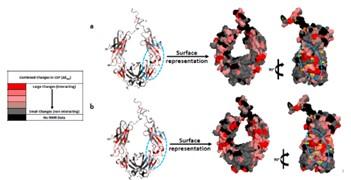
Recently, we performed these experiments with the Fc fragment of an IgG1 antibody and MM cation exchanging ligands (Capto MMC and Nuvia cPrime). The MM ligand binding sites on the Fc were concentrated in the hinge region and near the interface of the CH2 and CH3 domains which are relatively flexible regions of the Fc. Comparisons of the NMR results with protein surface properties suggested that the MM ligand binding regions were associated with overlapping regions of positive charge and hydrophobicity on the Fc surface.Recently, we performed these experiments with the Fc fragment of an IgG1 antibody and MM cation exchanging ligands (Capto MMC and Nuvia cPrime). The MM ligand binding sites on the Fc were concentrated in the hinge region and near the interface of the CH2 and CH3 domains which are relatively flexible regions of the Fc. Comparisons of the NMR results with protein surface properties suggested that the MM ligand binding regions were associated with overlapping regions of positive charge and hydrophobicity on the Fc surface.
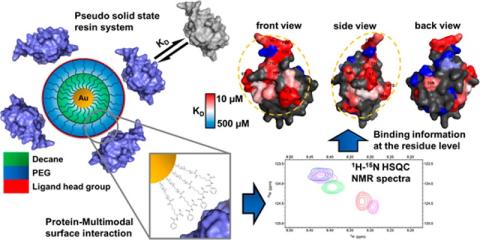
We have recently developed gold nanoparticles based system that mimics solid-state resins by presenting multimodal (MM) ligands on its surface and are also amenable to HSQC experiments. This enabled us to evaluate the binding regions on the protein surface for interacting with MM surfaces and make a more direct connection to the chromatographic condition. The results showed the preferred binding region on the protein surface as well as differences in the amino acid residues that interacted with two different MM ligand surfaces. This technique helped us to get fundamental insights into the nature of interactions in MM chromatographic systems.
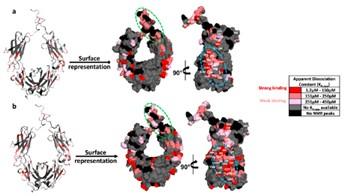
Similar experiments were also carried out with the Fc fragment of an IgG1. The results from the NMR experiments suggested that the binding of Fc to the MM-functionalized NPs resulted in stronger residue binding (μM affinities) as compared to the mM binding observed with the ligands in free solution. The stronger binding was possibly driven by the combination of cooperativity and avidity effects. The results showed that the binding sites were located on the CH2-CH3 interface and the hinge regions. While the binding in “Capto ligand” NPs were dominated by the hydrophobic interactions, in “Nuvia cPrime ligand” NPs, they were driven largely by the electrostatic interactions.
The effect of ligand density and salt concentrations were also investigated in this work.
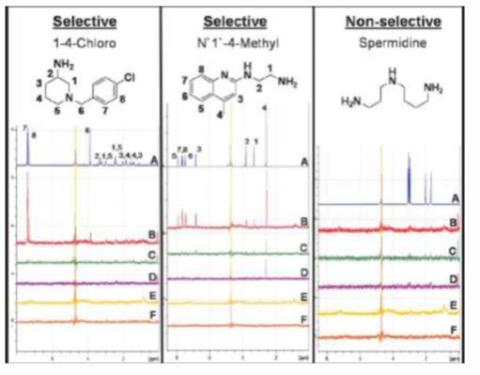
Saturation transfer difference (STD) NMR experiments are routinely used for examining protein-ligand binding in solution and this technique is often used as an initial screening technique for evaluating protein-ligand interactions and identifying the chemical moieties involved in binding. We have performed both HSQC and STD-NMR in the presence of fluid phase modifiers and/or displacers to probe the effect of these additives on the binding strength of different ligand moieties and elucidate a mechanism of action for these molecules.
Selected References:
- W.K. Chung, et al. PNAS (2010). Evaluation of protein adsorption and preferred binding regions in multimodal chromatography using NMR.
- M.A. Holstein, et al., J. Chromatogr. A (2012). Probing multimodal ligand binding regions on ubiquitin using nuclear magnetic resonance, chromatography, and molecular dynamics simulations.
- M.A. Holstein, et. al., Langmuir (2014). Effects of urea on selectivity and protein-ligand interactions in multimodal cation exchange chromatography.
- C.J. Morrison, et al. Biotechnol. Bioeng. (2009). Mechanistic Studies of Displacer-Protein Binding in Chemically Selective Displacement Systems Using NMR and MD Simulations.
- M.A. Holstein, et. al. Biotechnol. Bioeng. 109 (2012). Mobile phase modifier effects in multimodal cation exchange chromatography.