We are developing novel multimodal ligand libraries in order to better understand the relationship between ligand chemistry and preferred chromatographic behaviors such as separability and peak shape. These ligands are based on small molecule and short peptide frameworks. These ligands are synthesized and evaluated, the results of which are used to inspire the generation of future, enriched libraries.
Chromatography Retention
Chromatography Retention
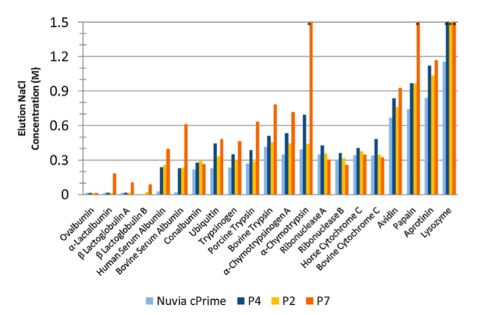
A multimodal cation exchange ligand library based on Nuvia cPrime was generated to better understand the relationship between ligand chemistries and chromatographic behavior.
Salt gradient elution experiments were run to show how substituting different hydrophobic groups altered the retention behavior of the ligand. This is seen in the figure comparing P4, P2, and P7 to Nuvia cPrime (Woo et al. Journal of Chromatography A 2015). In addition, varying the electrostatic group strength and linker length helps tease out the relationships between chemistry, stereochemistry, and retention behavior. Bettering our understanding of these relationships is critical to improving process development and novel ligand design.1
QSAR Modeling
QSAR Modeling
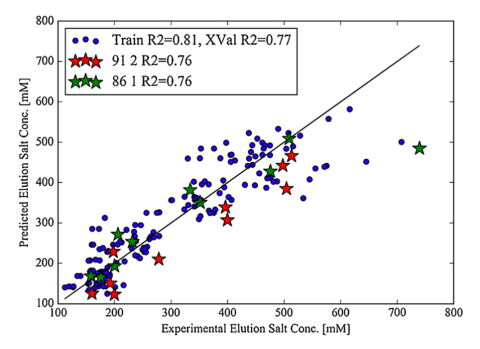
A multimodal anion exchange ligand library was generated to better understand the relationship between ligand chemistries and chromatographic behavior.
A large dataset of protein-ligand retention data was obtained in order to generate a partial least squares QSAR model. This model based on protein and ligand descriptors can accurately predict protein retention. In the figure, training data is seen in blue while test data is in green and red (Robinson et al. Journal of Chromatography A 2018). Using models like these can lead to the development of ligands with unique selectivities.1,2,3
Molecular Dynamics
Molecular Dynamics
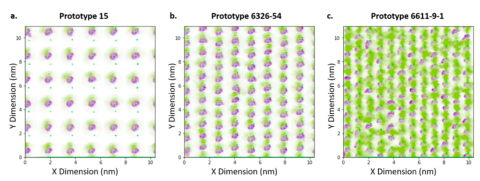
A multimodal ligand library was generated to better understand the relationship between ligand chemistries and chromatographic behavior.
Molecular dynamics simulations were employed to characterize the conformation preferences and the density distributions of the various ligands. The figure shows how the hydrophobic group impact relies on the ligand density (Koley et al. Journal of Chromatography A 2021). Molecular dynamics is another tool at our disposal to help generate better novel ligands.3
Separability and Orthogonality
Separability and Orthogonality
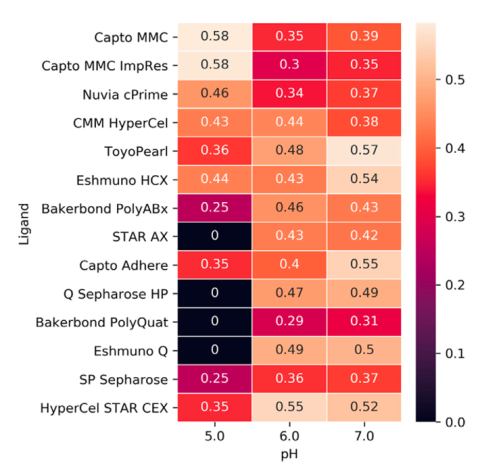
A methodology was developed to identify ligands that remove optimally orthogonal model proteins. Retention data is generated and is used in the calculation of separability and orthogonality metrics. These represent the ability of a ligand, or pairs of ligands, to separate a panel of diverse model proteins. The separabilities of various commercial ligands is shown in the figure (Bilodeau et al. Journal of Chromatography A 2020).
This is currently being used in the development of new ligands. Batch screening enables the generation of retention data on large, novel ligand libraries. These libraries can be screened rapidly, and this metric is one of many to aid us in identifying the most promising candidate ligands.4
Selected References:
(1) Woo, J. A., Chen, H., Snyder, M. A., Chai, Y., Frost, R. G., & Cramer, S. M. (2015). Defining the property space for chromatographic ligands from a homologous series of mixed-mode ligands. Journal of Chromatography A,1407, 58–68. https://doi.org/10.1016/j.chroma.2015.06.017
(2) Robinson, J., Snyder, M. A., Belisle, C., Liao, J. li, Chen, H., He, X., … Cramer, S. M. (2018). Investigating the impact of aromatic ring substitutions on selectivity for a multimodal anion exchange prototype library. Journal of Chromatography A, 1569, 101–109. https://doi.org/10.1016/j.chroma.2018.07.049
(3) Koley, S., Altern, S. H., Vats, M., Han, X., Jang, D., Snyder, M. A., … Cramer, S. M. (2021). Evaluation of guanidine-based multimodal anion exchangers for protein selectivity and orthogonality. Journal of Chromatography A, 1653, 462398. https://doi.org/10.1016/J.CHROMA.2021.462398
(4) Bilodeau, C. L., Vecchiarello, N. A., Altern, S., & Cramer, S. M. (2020). Quantifying Orthogonality and Separability: A Method for Optimizing Resin Selection and Design. Journal of Chromatography A, 1628, 461429. https://doi.org/10.1016/j.chroma.2020.461429